Food Safety Authority of Ireland becomes the first regulatory body to embrace next generation sequencing (NGS) to identify the complete DNA of food. NGS is no longer a buzz word, but reality now! But what exactly is it? This post introduces NGS to the uninitiated and talks about how it may be the perfect tool to address safety and quality threats related to authenticity, fraud, allergenicity and so on.
Let me break down NGS first.
All complex things start with something simple, in our case, let us begin with Sanger sequencing… or rather how DNA replicates (Excuse the nerd in me, my first degree was in biotechnology).
DNA replication goes as follows: a pre-existing strand of DNA is snipped, and the double strand is unzipped. Now a short piece of DNA called primer latches onto the strand, and DNA polymerase (an enzyme) adds nucleotides complementary to the parent strand and the strand elongates, and further termination begins.
Now Sanger used a ‘special nucleotide’ with a hydrogen atom instead of a hydroxyl group. This led to the strand to terminate whenever this nucleotide was added. With a few more chemicals and magic (science), the last ‘special nucleotide’ (*cough dideoxynucleotidetriphosphate) glows and we ‘read’ the sequence.
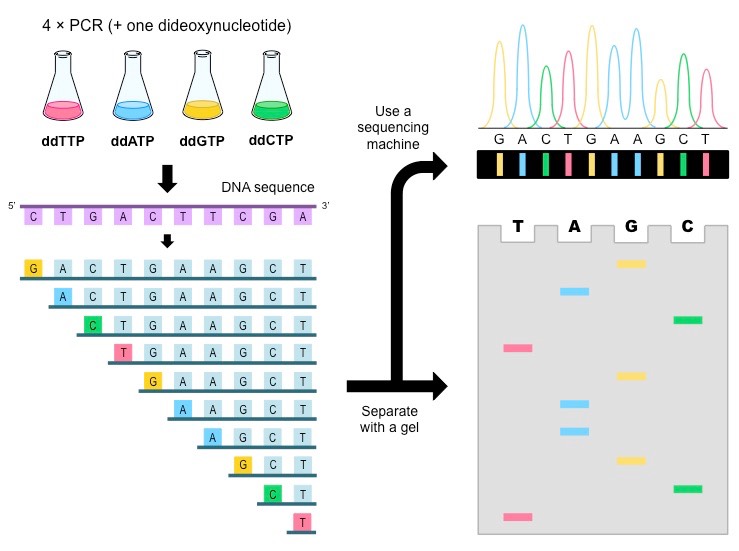
Well, NGS aka- Massively Parallel Sequencing is the overachieving sibling of Sanger sequencing and it can analyze millions of DNA data points in one shot !
It has three different applications such as
- Whole genome sequencing- Reads the whole sequence
- Shotgun Metabolomics- Imagine a crazy person with a shotgun, shooting everyone, same here. Only, an enzyme snips the DNA, We then ‘read’ these little pieces and fit the puzzle together using algorithms and other sciency stuff to understand the DNA.
- Targeted NGS- It specifically targets a particular marker in a genetic pool with high accuracy.
How can this help food?
NGS can help a wide variety of food microbiological investigations. It helps with source delineation in times of outbreaks. Let me give you an example.
Story time:
In 2010, Haiti was hit by a large earthquake resulting in losses of life and property, and weirdly- Cholera?? This is weird because Haiti never had a cholera outbreak, not in the past few decades and definitely not this massive- 9068 fatalities!
Where did it get it from? South America didn’t have cholera!
Traditional serotyping methods helped narrow the source down to- Afghanistan, Oman, Nepal, South Africa, Cameroon, India and Pakistan. Yeah, right. So. Not helpful. And then WGS came to our rescue, and the cholera came from aid workers sent from Nepal.
We don’t have this fancy technology for our routine investigations due to cost constraints. It is about this expensive.
Ireland revealed in Feb 2019 that NGS would be used for carrying out investigations pertaining to regulatory monitoring of food safety and quality.
WHAT ARE YOU EATING?
According to Dr. Pat Mahony, NGS allows to scan the entire DNA without suspecting what could be present! The FSAI investigated 45 plant-based products, of which 14 were selected for further investigation. They found undeclared mustard (a known allergen) at significant levels in one of the samples. They also found oregano to contain some undeclared plant species! Dr. Mahony says that it is important to corroborate this data with other findings and comments
“…The plan is that in the future, the FSAI will apply the same technology for the screening of meat, poultry and fish products,”
Dr. Mahony commenting on use of NGS in the future
Ireland is the only EU member states to have started using this technology, even the only country so far!
This is exciting news! I am sure that soon other countries will also have NGS as a regulatory benchmark.
Thanks for reading.
— Roo
Reference:
Orata, Fabini D., Paul S. Keim, and Yan Boucher. “The 2010 cholera outbreak in Haiti: how science solved a controversy.” PLoS pathogens 10, no. 4 (2014): e1003967.

Recent Comments